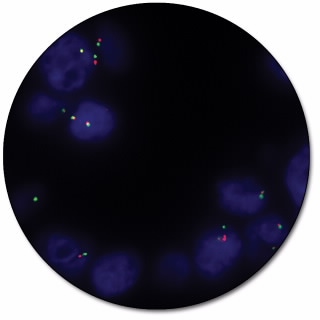
ALK IQFISH Break-Apart Probe
ALK IQFISH Break-Apart Probe (Dako Omnis) is intended for the detection of rearrangements involving the ALK gene by fluorescence in situ hybridization (FISH). The dual-color probe for Dako Omnis is recommended to be used on formalin-fixed, paraffin-embedded (FFPE) lung tissue specimens.
Break-apart probes consist of two child probes, designed to be on opposite sides of the translocation breakpoint for a given gene, each labeled with a different fluorochrome. These probes generate signals in normal cells that are closely matched in size and co-localized (two fusion signals). In case of a translocation event, the signals are ‘broken apart’ and no longer co-localize giving e.g. one red, one green and one fusion signal.
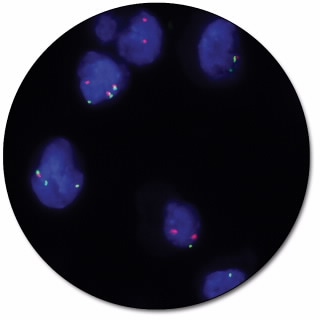
ROS1 IQFISH Break-Apart Probe
ROS1 IQFISH Break-Apart Probe (Dako Omnis) is intended for the detection of rearrangements involving the ROS1 gene by fluorescence in situ hybridization (FISH). The dual-color probe for Dako Omnis is recommended to be used on formalin-fixed, paraffin-embedded (FFPE) lung tissue specimens.
Break-apart probes consist of two child probes, designed to be on opposite sides of the translocation breakpoint for a given gene, each labeled with a different fluorochrome. These probes generate signals in normal cells that are closely matched in size and co-localized (two fusion signals). In case of a translocation event, the signals are ‘broken apart’ and no longer co-localize giving e.g. one red, one green and one fusion signal.
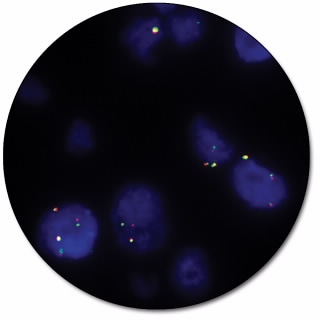
RET IQFISH Break-Apart Probe
RET IQFISH Break-Apart Probe (Dako Omnis) is intended for the detection of rearrangements involving the RET gene by fluorescence in situ hybridization (FISH). The dual-color probe for Dako Omnis is recommended to be used on formalin-fixed, paraffin-embedded (FFPE) lung tissue specimens.
Break-apart probes consist of two child probes, designed to be on opposite sides of the translocation breakpoint for a given gene, each labeled with a different fluorochrome. These probes generate signals in normal cells that are closely matched in size and co-localized (two fusion signals). In case of a translocation event, the signals are ‘broken apart’ and no longer co-localize giving e.g. one red, one green and one fusion signal.
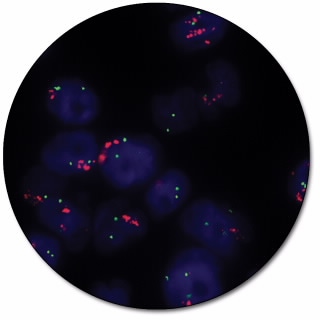
MET IQFISH Probe with CEP7
MET IQFISH Probe with CEP7 (Dako Omnis) is intended for the detection of the MET gene amplification by fluorescence in situ hybridization (FISH). The dual-color probe for Dako Omnis is recommended to be used on formalin-fixed, paraffin-embedded (FFPE) lung tissue specimens.
In normal diploid cells in metaphase or interphase, the MET IQFISH Probe with CEP7 generates two orange-red signals (MET) and two green signals (chromosome 7). In cells with a gain or amplification of the MET gene or of chromosome 7, the number of signals per cell will be greater.
